論文タイトル: Iterative energy-based projection on a normal data manifold for anomaly localization
注:qiitaから移転
論文リンク
ICLR 2020: https://openreview.net/forum?id=HJx81ySKwr
解説スライド
コード
データはMVTecADを使った
まずはデータローダの作成
# data loader
import os
import numpy as np
from PIL import Image
import torch
from torch.utils import data
from torchvision import transforms as T
import torch.nn as nn
import torch.optim as optim
from torch.nn import functional as F
import matplotlib.pyplot as plt
class MVTecAD(data.Dataset):
"""Dataset class for the MVTecAD dataset."""
def __init__(self, image_dir, transform):
"""Initialize and preprocess the MVTecAD dataset."""
self.image_dir = image_dir
self.transform = transform
def __getitem__(self, index):
"""Return one image"""
filename = "{:03}.png".format(index)
image = Image.open(os.path.join(self.image_dir, filename))
return self.transform(image)
def __len__(self):
"""Return the number of images."""
return len(os.listdir(self.image_dir))
def return_MVTecAD_loader(image_dir, batch_size=256, train=True):
"""Build and return a data loader."""
transform = []
transform.append(T.Resize((512, 512)))
transform.append(T.RandomCrop((128,128)))
transform.append(T.RandomHorizontalFlip(p=0.5))
transform.append(T.RandomVerticalFlip(p=0.5))
transform.append(T.ToTensor())
transform = T.Compose(transform)
dataset = MVTecAD(image_dir, transform)
data_loader = data.DataLoader(dataset=dataset,
batch_size=batch_size,
shuffle=train)
return data_loaderデータはいろんな種類のものがあるがgridデータのみを使用した
train_loader = return_MVTecAD_loader("./mvtec_anomaly_detection/grid/train/good/")
device = torch.device("cuda" if torch.cuda.is_available() else "cpu")
print(device)
seed = 42
out_dir = './logs'
if not os.path.exists(out_dir):
os.mkdir(out_dir)
torch.manual_seed(seed)
if torch.cuda.is_available():
torch.cuda.manual_seed(seed)VAEのモデルはこんな感じで作った.
class VAE(nn.Module):
def __init__(self, z_dim=128):
super(VAE, self).__init__()
# encode
self.conv_e = nn.Sequential(
nn.Conv2d(1, 32, kernel_size=4, stride=2, padding=1), # 128 ⇒ 64
nn.BatchNorm2d(32),
nn.LeakyReLU(0.2),
nn.Conv2d(32, 64, kernel_size=4, stride=2, padding=1), # 64 ⇒ 32
nn.BatchNorm2d(64),
nn.LeakyReLU(0.2),
nn.Conv2d(64, 128, kernel_size=4, stride=2, padding=1), # 32 ⇒ 16
nn.BatchNorm2d(128),
nn.LeakyReLU(0.2),
)
self.fc_e = nn.Sequential(
nn.Linear(128 * 16 * 16, 1024),
nn.BatchNorm1d(1024),
nn.LeakyReLU(0.2),
nn.Linear(1024, z_dim*2),
)
# decode
self.fc_d = nn.Sequential(
nn.Linear(z_dim, 1024),
nn.BatchNorm1d(1024),
nn.LeakyReLU(0.2),
nn.Linear(1024, 128 * 16 * 16),
nn.LeakyReLU(0.2)
)
self.conv_d = nn.Sequential(
nn.ConvTranspose2d(128, 64, kernel_size=4, stride=2, padding=1),
nn.BatchNorm2d(64),
nn.LeakyReLU(0.2),
nn.ConvTranspose2d(64, 32, kernel_size=4, stride=2, padding=1),
nn.BatchNorm2d(32),
nn.LeakyReLU(0.2),
nn.ConvTranspose2d(32, 1, kernel_size=4, stride=2, padding=1),
nn.Sigmoid()
)
self.z_dim = z_dim
def encode(self, input):
x = self.conv_e(input)
x = x.view(-1, 128*16*16)
x = self.fc_e(x)
return x[:, :self.z_dim], x[:, self.z_dim:]
def reparameterize(self, mu, logvar):
if self.training:
std = logvar.mul(0.5).exp_()
eps = std.new(std.size()).normal_()
return eps.mul(std).add_(mu)
else:
return mu
def decode(self, z):
h = self.fc_d(z)
h = h.view(-1, 128, 16, 16)
return self.conv_d(h)
def forward(self, x):
mu, logvar = self.encode(x)
z = self.reparameterize(mu, logvar)
self.mu = mu
self.logvar = logvar
return self.decode(z)
model = VAE(z_dim=512).to(device)訓練
def loss_function(recon_x, x, mu, logvar):
recon = F.binary_cross_entropy(recon_x, x, reduction='sum')
kld = -0.5 * torch.sum(1 + logvar - mu.pow(2) - logvar.exp())
return recon + kld
def train(epoch):
model.train()
train_loss = 0
for batch_idx, data in enumerate(train_loader):
data = data.to(device)
optimizer.zero_grad()
recon_batch = model(data)
loss = loss_function(recon_batch, data, model.mu, model.logvar)
loss.backward()
train_loss += loss.item()
optimizer.step()
train_loss /= len(train_loader.dataset)
return train_loss
# gif作成用
def iterative_plot(x_t, j):
plt.figure(figsize=(15, 4))
for i in range(10):
plt.subplot(1, 10, i+1)
plt.xticks([])
plt.yticks([])
plt.imshow(x_t[i][0], cmap=plt.cm.gray)
plt.subplots_adjust(wspace=0., hspace=0.)
plt.savefig("./results/{}.png".format(j))
plt.show()500epochくらい回した
optimizer = optim.Adam(model.parameters(), lr=5e-4)
num_epochs = 500
for epoch in range(num_epochs):
loss = train(epoch)
print('epoch [{}/{}], train loss: {:.4f}'.format(
epoch + 1,
num_epochs,
loss))推論
model.eval()
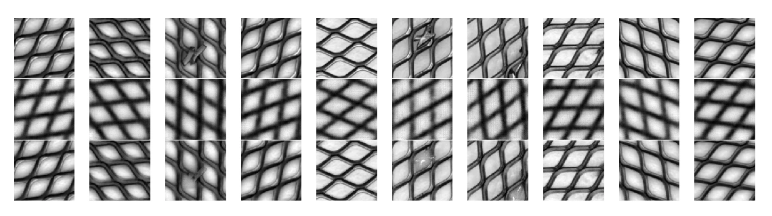
test_loader = return_MVTecAD_loader("./mvtec_anomaly_detection/grid/test/metal_contamination/", batch_size=10, train=False)まずは単純なVAEのとき,傷が消えてるものの,ぼやけていることを確認する
x_0 = iter(test_loader).next()
model.eval()
with torch.no_grad():
x_vae = model(x_0.to(device)).detach().cpu().numpy()上が元画像,下が再構成.

次に提案手法について
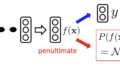
$$
E(x_t) = L_r(x_t) + \lambda ||x_t-x_0||_1
$$
$$
x_{t+1} = x_t – \alpha\cdot(\nabla_xE(x_t)\odot (x_t – f_{VAE}(x_t))^2)
$$
上記式を実装するだけ
# ハイパラ
alpha = 0.05
lamda = 1
x_0 = x_0.to(device).clone().detach().requires_grad_(True)
recon_x = model(x_0).detach()
loss = F.binary_cross_entropy(x_0, recon_x, reduction='sum')
loss.backward(retain_graph=True)
x_grad = x_0.grad.data
x_t = x_0 - alpha * x_grad * (x_0 - recon_x) ** 2
for i in range(15):
recon_x = model(x_t).detach()
loss = F.binary_cross_entropy(x_t, recon_x, reduction='sum') + lamda * torch.abs(x_t - x_0).sum()
loss.backward(retain_graph=True)
x_grad = x_0.grad.data
x_t = x_t - eps * x_grad * (x_t - recon_x) ** 2
iterative_plot(x_t.detach().cpu().numpy(), i)gifなのでしばらく眺めてください

画像が割と鮮明なまま,異常箇所のみが消えていくことが確認した
もう一度VAEと比較(上段: テスト画像,中段: VAEによる再構成,下段: 提案手法による再構成)


コメント